Characterizing the alterations involved in premalignant progression
To truly identify alterations that may be important for progression, testing samples with precise knowledge of the histologic diagnosis and outcome (progressor vs. non-progressor) must be utilized. To this end, the lab has predominately focused on formalin fixed paraffin embedded samples and have used laser capture microdissection to cut out the exact cell population that is to be tested and have begun expanding into single cell approaches. Ongoing and future projects are focused on 1) understanding the genomic events around the transition from high grade dysplasia into very early cancers (before the cancer has a chance to develop additional complex alterations) through whole genome and single cell DNA sequencing. 2) Understanding the events underlying resistance and recurrence post treatment. 3) Expanding beyond genomics to explore how epigenetic alterations evolve through the progression process.
Functional interrogation of identified alterations
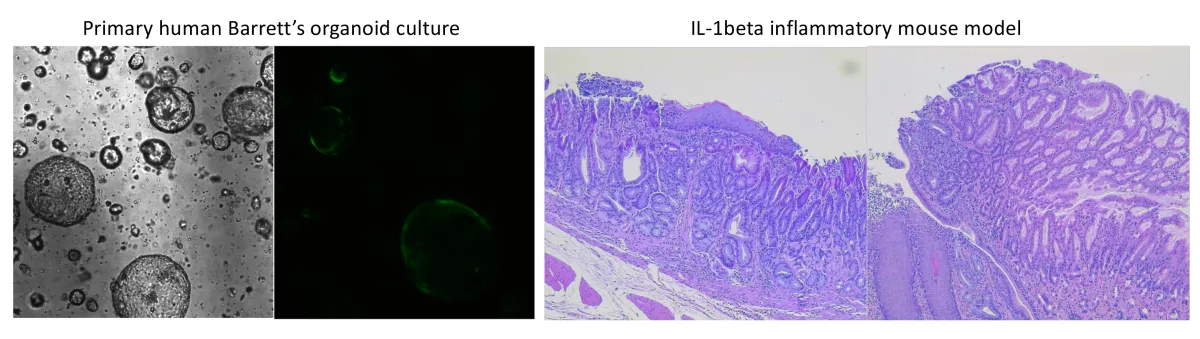
To truly understand what the consequence of the identified alterations are, we are using a combination of organoids derived from human Barrett’s biopsies and a murine model of metaplasia/esophagogastric adenocarcinoma. Using both CRISPR-Cas9 and other genomic manipulation techniques we are beginning to introduce genomic alterations into these cells and to determine their functional consequences.
Understanding how the immune microenvironment evolves,
interacts with genomic and other alterations, and influences progression
While the alterations that occur within the Barrett’s epithelial cells are critical to progression, Barrett’s arises in a hostile environment with chronic exposure to bile and acid reflux as well as an intense inflammatory background. These non-intrinsic factors are also likely critical to the process of progression. Our goal is to first understand what type of cells are present and how these populations change over time. We then want to understand how the cells of the microenvironment may interact with the different alterations that develop within the Barrett’s epithelial cells. We are utilizing a mixture of multiplex immunostaining and spatial transcriptomics of human tissue as well as both in vitro co-cultures and an inflammatory murine model.
Development of biomarkers and models of progression risk
As a translational research lab, the end goal of all of our work is to improve patient outcome. Using the information we have gained through previous studies and adapting cutting edge sequencing technologies and analysis to work in a clinical setting, we are beginning to test a biomarker panel developed for progression risk stratification in patients with Barrett’s esophagus. This is being performed on an extensive and well annotated collection of clinical Barrett’s biopsies, giving an unprecedented opportunity to understand the differences between patients with Barrett’s esophagus who go on to progress to advanced disease and those who harbor indolent Barrett’s esophagus who have stable disease over many years. Moving beyond retrospective archived samples, we are beginning to test other sampling techniques (such endoscopic brushes and balloon/sponge devices) to compare their ability to identify biomarkers of interests compared to the standard of care (random four quadrant biopsies).